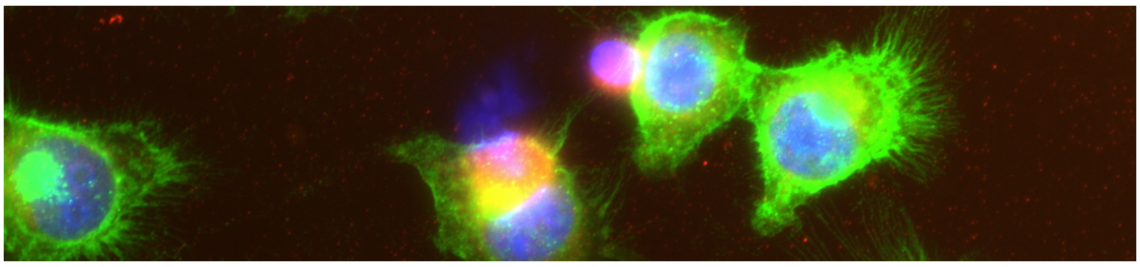
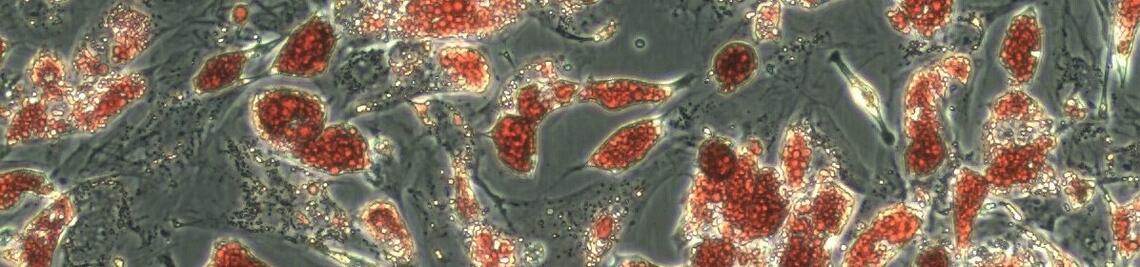
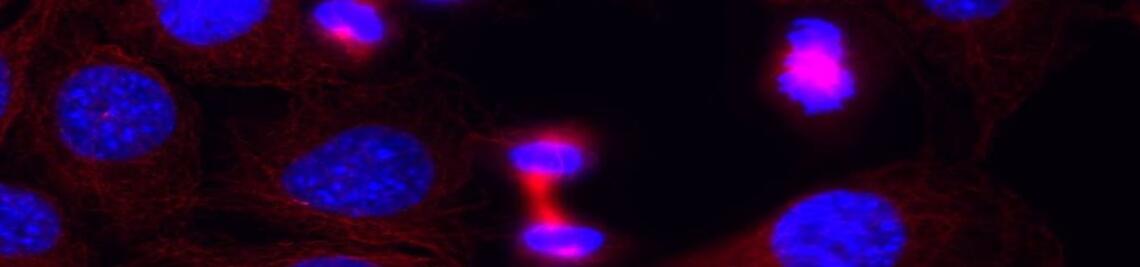
Workshops
You always wanted to learn about optogenetics, single-cell transcriptomics, and personalized oncology? Or you would like to know how professional pipetting or the development of next generation multispecific antibodies works? Or what it's like to conduct research with in vitro models as an alternative to animal testing?
Here, all available workshops provided by research groups of the Julius Maximilian University are listed, categorized into the research focuses of Würzburg: cardiovascular diseases, neurosciences, tumor research, infection biology & immunology.
If you are interested in participating in one ore two of them, you can state so in the registration form. You can choose one workshop for the morning session and one for the afternoon session. Please notice, that the registration for each workshop will be closed, as soon as the max. number of participants is reached. Requirements for participation are indicated if necessary.
Morning Workshops
[A1.1] The Role of Next-Generation Sequencing in Personalized Oncology
Organizer: Dr. Silke Appenzeller
Institute/Department: Comprehensive Cancer Center Mainfranken
Abstract: The development of molecularly targeted therapies and immunotherapies has led to major advances in the treatment of patients with cancer. By means of Next Generation Sequencing (NGS) techniques, potentially targetable and/or tumor-driving alterations and resistance mechanisms can be detected. Furthermore, the genetic alterations can contribute to the characterization of the tumor type. The results can serve as a basis for personalized therapy decisions. In this workshop, the opportunities and limitations of NGS methods used for tumor diagnostics will be presented.
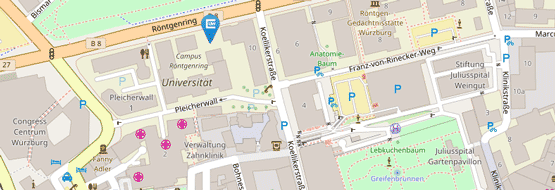
Location: Seminar room 01.006, RVZ, Josef-Schneider-Straße 2, Bulding D15
Max. number of participants: 10
Target group: Master.
Remark: Participants should bring their own laptop. It may be necessary to install some freely available tools for data analysis.
[A1.2] Single cell transcriptomics - how does it work?

Organizer: Dr. Andy Shing Fung Chan
Institute/Department: Max Planck Research Group for Systems Immunology (WÜSI)
Abstract: Single cell transcriptomic technologies have revolutionized biological research by enabling high-resolution analysis of individual cells. They provide insights into cellular heterogeneity, states, and dynamics, leading to a deeper understanding of complex biological systems. Learning these technologies equips researchers with powerful tools to unravel disease mechanisms, discover novel cell types, identify therapeutic targets, and advance precision medicine. In this workshop, participants will be exposed to knowledge of basic principles and methods of single cell transcriptomics, and to perform simple analysis of the generated single cell data.
Location: 3rd Floor, Institut für Systemimmunology, Building E6, Versbacher Str. 9, 97078 Würzburg
Max. number of participants: 10
Target group: Master. (Basic knowledge about molecular biology technique, such as PCR, is required. Basic programming skills are optional.)
Remark: Please bring your own laptop. Ideally, "R" and "R Studio" are already installed.
[A1.3] Biomedical research using organoid technology

Organizer: Dr. Kai Kretzschmar
Institute/Department: MSNZ Würzburg
Abstract: Organoid technology has emerged as a powerful tool for biomedical research. Organoids are three-dimensional clusters of cells derived from self-organising stem cells that resemble key features of their tissue of origin. In the laboratory, we use organoid technology to study epithelial homeostasis and tumourigenesis. In the workshop, you will learn more about the methodology and get hands-on experience of working with adult stem cell-derived epithelial organoid cultures.
Location: MSNZ Würzburg, Institute for Virology and Immunobiology, Building E5, 2nd floor, Room 316, Versbacher Str. 7, 97087 Würzburg
Max. number of participants: 4
Target group: Bachelor & Master. (You should have experience with cell culture.)
[A1.4] Analysis of the complexity of the tumor microenvironment using single-cell RNA sequencing

Organizer: Dr. Angela Riedel (with Moutaz Helal, PhD student)
Institute/Department: MSNZ Würzburg
Abstract: In this course, we will explore various methods for analyzing single-cell data using the R programming language. The workshop will begin with an introduction to single-cell sequencing technologies, and how it is used to understand tumor entities. We will then dive into the basics of R programming and data manipulation in R. Once we have covered the basics, we will get to know “Seurat” one of the most widely used tools to analyze omics data. Then, we will move on to more advanced topics, such as data normalization, dimensional reduction, clustering, cell type assignment, and differential expression analysis.
Location: Insitute of Virology and Immunology, Versbacher Str. 7, 97078 Würzburg, Building E5
Max. number of participants: 6
Target group: Master. (You should have basic knowledge of RNA sequencing techniques. Understanding of tumor biology and cell lineages may be advantageous.)
[A1.5] Single Cell Biology Introduction

Organizer: Prof. Dr. Emmanuel Saliba
Organization: Helmholtz Institute for RNA-based infection research
Abstract: The basis of lung injury induced by SARS-CoV-2 remains poorly understood. In the severe COVID-19 cases, patients develop COVID-19-induced Acute Respiratory Distress Syndrome (ARDS) with prolonged respiratory failure and high mortality. To comprehend the pathomechanisms of lung damage, we analyzed pulmonary immune responses and lung pathology in patients with COVID-19 ARDS using functional single-cell genomics, immunohistology, and electron microscopy. Phenotypically, we observed an accumulation of CD163-expressing monocyte-derived macrophages that acquired a profibrotic transcriptional phenotype during COVID-19 ARDS. COVID-19 ARDS was associated with clinical, radiographic, and histopathological characteristics of pulmonary fibrosis. Exposure of human monocytes to SARS-CoV-2, but not influenza A or viral RNA analogs, was sufficient to induce a similar profibrotic phenotype in vitro. Yet, the role of macrophages remains intriguing, and we will discuss whether they are friends or foes.
Location: Seminar room 01.002, RVZ, Josef-Schneider-Straße 2, Bulding D15
Max. number of participants: 10
Target group: Master.
Remark: Please bring your own laptop with "R" and "R Studio" already installed.
[A1.6] Basics of pipetting - what influences pipetting accuracy?

Organizer: Dr. Hans von Besser
Institute/Department: Eppendorf Vertrieb Deutschland GmbH
Abstract: The technological basis of pipetting and dispensing will be presented. Limits of air cushion pipettes (eg. Eppendorf Research® plus) and the advantages of electronic pipettes (Eppendorf Xplorer®) and positive displacement pipettes (Multipette®) will be figured out. Several factors (environmental, physical) have a significant impact on the accuracy of the pipetting results. Best practice of liquid handling is important in many applications like e.g. qPCR. Several practical pipetting tasks will give you the opportunity to compare the advantages of air cushion pipettes vs. positive displacement pipettes.
Location: Seminar room 01.004, RVZ, Josef-Schneider-Straße 2, Bulding D15
Max. number of participants: 12
Target group: Bachelor. (You should already have some experience with pipetting.)
[A1.7] Examination of the human sensory system

Organizer: Prof. Claudia Sommer / Prof. Heike Rittner
Institute/Department: Department of Neurology / Department of Anesthesiology
Abstract: The Quantitative Sensory Testing (QST) workshop is a practical and immersive experience that provides participants with the knowledge and skills to objectively measure and characterize sensory experiences. Through hands-on training, attendees will learn the principles, techniques, and applications of QST, including stimulus manipulation, data collection, analysis, and interpretation. Join us to gain a deeper understanding of sensory perception and discover the applications, and consequent implications, of using QST in clinical practice and research, as an important tool for diagnosing sensory disorders and optimizing pain control.
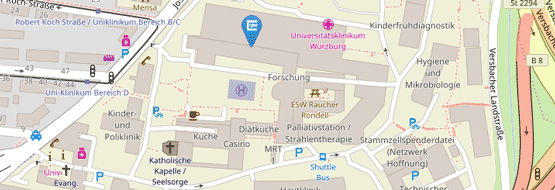
Location: Room 1802A and 1631, Kopfklinik B1, Josef-Schneider-Straße 11, 97080 Würzburg
Max. number of participants: 8
Target group: Master. (You should be interested in clinically oriented or translational research.)
[A1.8] Optogenetics

Organizer: Prof. Dr. Philip Tovote
Institute/Department: Insitute of Clinical Neurobiology, Defense Circuits Lab
Abstract: The goal of this course is to understand how optogenetics can be used to investigate specific neuronal circuits and their associated behavioral response. Students will get a comprehensive introduction into optogenetics and will be guided through the process of designing an experiment. By activating channelrhodopsin in motorneurons of transgenic Drosophila melanogaster, we will assess the function of these neurons on animal locomotion. Data will be analyzed using a custom written software.
Location: Lecture Hall (lower floor), Institute of Clinical Neurobiology, Versbacherstraße 5, Building E4
Max. number of participants: 10
Target group: Master. (You should have basic knowledge in neuroscience.)
Remark: Participants should bring their own labcoat.
[A1.9] Ion channels at work

Organizer: Prof. Dr. Carmen Villmann
Institute/Department: Institute for Clinical Neurobiology
Abstract: The workshop is about the introduction of electrophysiological techniques to characterize ion channels. First, information is given about the use of electrophysiological recording techniques to help identifying molecular pathomechanisms in various forms of neurological diseases. The practical part will include demonstration of recordings from transfected cells and primary neurons. Student will have the chance to do hands-on electrophysiology by trying patch a cell on their own.
Location: Seminar Room, Institute for Clinical Neurobiology E4, Versbacherstraße 5, 97080 Würzburg
Max. number of participants: 8
Target group: Bachelor & Master.
[A2.1] Single-cell imaging of cell cycle regulation and DNA damage in mammalian cells

Organizer: Prof. Dr. Stefan Gaubatz
Institute/Department: Biocenter, Department of Biochemistry and Cell Biology
Abstract: An in-depth understanding of cell cycle regulation holds significant importance in the fields of cancer biology and regenerative medicine. This workshop aims to provide participants with the knowledge and skills to utilize fluorescence microscopy as a powerful alternative to flow cytometry, enabling accurate identification of different cell cycle phases in both fixed and live cells. Through the utilization of various markers, such as fluorescence dyes or antibodies, participants will learn how to distinguish cells in G1, S-phase, G2, and mitosis. Additionally, we will explore the quantification of cell cycle stage-specific DNA damage, replication origin licensing, and cyclin-dependent kinase activity.
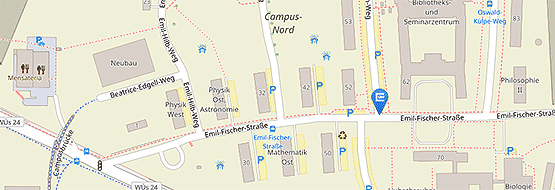
Location: Biocenter, Am Hubland, 97074 Würzburg
Max. number of participants: 6-8
Target group: Bachelor & Master. (You should be interested in mammalian cell cycle regulation and its dysregulation in cancer.)
[A2.2] In vitro models as an alternative to animal testing

Organizer: Dr. Florian Groeber-Becker, Fraunhofer ISC
Institute/Department: Translationszentrum
Abstract: Tissue engeneering is at the forefront of biomedical research, offering promising solutions for organ transplantation, disease modeling, and drug testing. In vitro models play a pivotal role in mimicking the complex physiological environments of tissues and organs, enabling the study of disease mechanisms and drug responses. This workshop is designed to provide biomedical students with a comprehensive overview of the latest developments in tissue engineering, with a specific emphasis on in vitro models as an alternative to animal models including organoid culture, lab automation, bioreactor design, and material science.
Location: Röntgenring 12, Würzburg
Max. number of participants: 12
Target group: Master.
[A2.3] Preclinical development of next generation multispecific antibodies

Organizer: Dr. Isabell Lang
Institute/Department: Department of Internal Medicine II, Division of Molecular Internal Medicine
Abstract: We give insights into the preclinical development and the requirements for next-generation multispecific antibodies and how they can be generated via genetic engineering. The practical part of the workshop is divided into several sections: production of the antibodies in cell culture, analyzing the antibody binding behaviour (cellular binding studies) and demonstrating the specific activity of the multispecific antibodies via ELISA.
Location: Division of Molecular Internal Medicine, Auverahaus, 1.OG, AG Wajant, Grombühlstraße 12
Max. number of participants: 10
Target group: Bachelor & Master. (Cell culture experience is optional.)
Afternoon Workshops
[B1.1] Analysis of neuronal tissue with bioimages (microscopy images)

Organizer: Professor Dr. Robert Blum
Institute/Department: Department of Neurology
Abstract: Modern microscopy enables researchers to capture images that describe cellular and molecular features in biological samples at an unprecedented scale. One of the most frequently used imaging methods is fluorescent labeling of macromolecules. To test a hypothesis, image features have to be interpreted and analyzed quantitatively, a process known as bioimage analysis. In this workshop, we will introduce properties of bioimages and will demonstrate and discuss recent developments for computer-aided, large-scale analysis of microscopy data.
Description: We will provide bioimages and will introduce how microscopy images can be analyzed. We will discuss manual as well as modern computational options (e.g. Deep Learning-based bioimage analysis).
Location: Seminar room 00.048, RVZ, Josef-Schneider-Straße 2, Bulding D15
Max. number of participants: 12
Target group: Bachelor & Master. A certain experience or knowledge of imaging, microscopy, or fluorescence labeling of cells or tissue is required.
Remark: Participants should bring their laptop with a Fiji (ImageJ) installation.
[B1.2] Investigating dynamics and interactions of viral RNAs at single molecule level

Organizer: Jun. Prof. Dr. Neva Caliskan
Institute/Department: Helmholtz Institute for RNA based Infection Research, Recoding Mechanisms in Infections
Abstract: Viral RNAs, like those studied in the Caliskan lab, exhibit dynamic conformations and folding behaviors. These characteristics influence their interactions with host and viral encoded factors, rendering them potential candidates for RNA-based antiviral therapeutics. New technologies in RNA research enables precise measurement and mapping of RNA structure and RNA-factor interactions at single molecule resolution in highly controlled environments. In this interactive workshop, students will explore RNA folding dynamics of fluorescent molecules using high-resolution optical tweezers.
Location: Seminar room 02.016 + 02.017, Josef Schneider Straße 2 D15, 97080 Würzburg
Max. number of participants: 5-6
Target group: Bachelor & Master.
[B1.3] Single Cell Genomics Data Analysis

Organizer: Dr. Abdelrahman Mahmoud
Institute/Department: Max Planck Research Group for Systems Immunology (WÜSI) / Medical Systems Biology Department
Abstract: The discovery of the cells changed our perspective on the underlying structure of tissues and organs. We began to understand that cellular alterations could have a causal link to diseases, and we started to catalog human diseases accordingly. Now, new high-throughput technologies enable us to unravel the underlying genetic and epigenetic regulation of the biological systems, coupled with imaging and spatial information at the “single-cell” resolution. These technologies generated a huge and high-dimensional data which gave us compelling challenges to develop computational approaches for data analyses and integration. This workshop will be an exciting journey to understand the data science of single-cell genomics. You will learn about the state of the art computational methods, recently developed in our field. The workshop will help you to gain the know-how skills and knowledge to be part of this new and revolutionary understanding of the biological systems in health and disease.
Location: Seminar room 01.006, RVZ, Josef-Schneider-Straße 2, Bulding D15, 97080 Würzburg
Max. number of participants: 50
Target group: Bachelor & Master.
Remark: Participants should bring their own laptop. Knowledge of a programming language (R or Python) is required.
[B1.4] Biomedical research using organoid technology

Organizer: Dr. Kai Kretzschmar
Institute/Department: MSNZ Würzburg
Abstract: Organoid technology has emerged as a powerful tool for biomedical research. Organoids are three-dimensional clusters of cells derived from self-organising stem cells that resemble key features of their tissue of origin. In the laboratory, we use organoid technology to study epithelial homeostasis and tumourigenesis. In the workshop, you will learn more about the methodology and get hands-on experience of working with adult stem cell-derived epithelial organoid cultures.
Location: MSNZ Würzburg, Institute for Virology and Immunobiology, Building E5, 2nd floor, Room 316, Versbacher Str. 7, 97087 Würzburg
Max. number of participants: 4
Target group: Bachelor & Master. (You should have experience with cell culture.)
[B1.5] Metabolism and excitation-contraction coupling in the normal and failing heart

Organizer: Prof. Dr. med. Christoph Maack, Dr. rer. Nat. Michael Kohlhaas, Dr. rer. Nat. Alexander G. Nickel
Institute/Department: Department of Translational Research, Comprehensive Heart Failure Center (CHFC), University Clinic Würzburg
Abstract: In heart failure, major defects are dysregulated calcium handling, energetic deficit and oxidative stress in cardiac myocytes. Our group works on the mechanisms how these defects link together, since defects in calcium handling as well as mechanical workload of the failing heart induces oxidative stress that drives cardiac remodeling and dysfunction. In our workshop, we explain and demonstrate the methods with which we measure calcium handling, mitochondrial energetics and reactive oxygen species formation in isolated cardiac myocytes or isolated mitochondria.
Location: Room A15.0.301 + A15.0.302, in the DZHI (Am Schwazenberg 15, Building A15)
Max. number of participants: 8
Target group: Bachelor & Master.
[B1.6] Immunophenotpying myocardial immune cells

Organizer: Dr. Gustavo Ramos & Dr. Clement Cochain
Institute/Department: Comprehensive Heart Failure Center
Abstract: The Workshop will discuss protocols to purify immune cells from solid tissues (e.g. heart) for downstream immunophenotyping applications, such as flow cytometry, cell sorting, and single-cell-sequencing. After a brief theoretical introduction, the speakers will analyze together with the students previously acquired flow cytometry samples and existing single-cell sequencing datasets. Though the discussions will primarily focus on myocardial cell biology, similar principles can be extrapolated to other solid tissues
Location: Room A15.0.306, in the DZHI (Am Schwazenberg 15, Building A15)
Max. number of participants: 15
Target group: Bachelor & Master.
[B1.7] Single Cell Biology Introduction

Organizer: Prof. Dr. Emmanuel Saliba
Organization: Helmholtz Institute for RNA-based infection research
Abstract: The basis of lung injury induced by SARS-CoV-2 remains poorly understood. In the severe COVID-19 cases, patients develop COVID-19-induced Acute Respiratory Distress Syndrome (ARDS) with prolonged respiratory failure and high mortality. To comprehend the pathomechanisms of lung damage, we analyzed pulmonary immune responses and lung pathology in patients with COVID-19 ARDS using functional single-cell genomics, immunohistology, and electron microscopy. Phenotypically, we observed an accumulation of CD163-expressing monocyte-derived macrophages that acquired a profibrotic transcriptional phenotype during COVID-19 ARDS. COVID-19 ARDS was associated with clinical, radiographic, and histopathological characteristics of pulmonary fibrosis. Exposure of human monocytes to SARS-CoV-2, but not influenza A or viral RNA analogs, was sufficient to induce a similar profibrotic phenotype in vitro. Yet, the role of macrophages remains intriguing, and we will discuss whether they are friends or foes.
Location: Seminar room 01.002, RVZ, Josef-Schneider-Straße 2, Bulding D15
Max. number of participants: 10
Target group: Master.
Remark: Please bring your own laptop with "R" and "R Studio" already installed.
[B1.8] Single cell phenotyping using flow cytometry to decipher hematological disorders

Organizer: Prof. Dr. Harald Schulze
Institute/Department: University Hospital Würzburg, Institute of Experimental Biomedicine, Chair I
Description: We will present translational methods that can be used to study human diseases applying mouse models.
Abstract: Hematopoietic stem cells, the progenitors of all blood cells, can differentiate to megakaryocytes (MKs) that produce blood platelets. Bone marrow MKs are rare cells and not easy to visualize. We will detect MKs in situ by confocal laser scanning microscopy. Additionally, we will characterize platelet receptor expression and function, including the kinetics of dense granule release, by flow cytometry. Finally, we will demonstrate data analysis using different modules of the software FlowJo.
Location: Josef-Schneider-Str. 2 D16, 3rd floor
Max. number of participants: 8
Target group: Bachelor. (You should be interested in the interface of basic science and clinical diagnostics)
[B1.9] Basics of pipetting - what influences pipetting accuracy?

Organizer: Dr. Hans von Besser
Institute/Department: Eppendorf Vertrieb Deutschland GmbH
Abstract: The technological basis of pipetting and dispensing will be presented. Limits of air cushion pipettes (eg. Eppendorf Research® plus) and the advantages of electronic pipettes (Eppendorf Xplorer®) and positive displacement pipettes (Multipette®) will be figured out. Several factors (environmental, physical) have a significant impact on the accuracy of the pipetting results. Best practice of liquid handling is important in many applications like e.g. qPCR. Several practical pipetting tasks will give you the opportunity to compare the advantages of air cushion pipettes vs. positive displacement pipettes.
Location: Seminar room 01.004, RVZ, Josef-Schneider-Straße 2, Bulding D15
Max. number of participants: 12
Target group: Bachelor. (You should already have some experience with pipetting.)
[B2.1] Single-cell imaging of cell cycle regulation and DNA damage in mammalian cells

Organizer: Prof. Dr. Stefan Gaubatz
Institute/Department: Biocenter, Department of Biochemistry and Cell Biology
Abstract: An in-depth understanding of cell cycle regulation holds significant importance in the fields of cancer biology and regenerative medicine. This workshop aims to provide participants with the knowledge and skills to utilize fluorescence microscopy as a powerful alternative to flow cytometry, enabling accurate identification of different cell cycle phases in both fixed and live cells. Through the utilization of various markers, such as fluorescence dyes or antibodies, participants will learn how to distinguish cells in G1, S-phase, G2, and mitosis. Additionally, we will explore the quantification of cell cycle stage-specific DNA damage, replication origin licensing, and cyclin-dependent kinase activity.
Location: Biocenter, Am Hubland, 97074 Würzburg
Max. number of participants: 6-8
Target group: Bachelor & Master. (You should be interested in mammalian cell cycle regulation and its dysregulation in cancer.)