New review article on the control of ribosome biogenesis by nucleolar long non-coding (lnc)RNA
22.07.2021Victoria Mamontova, PhD student in Kaspar Burger's group, has published a review article on how RNA polymerase II influences nucleolar structure and function.
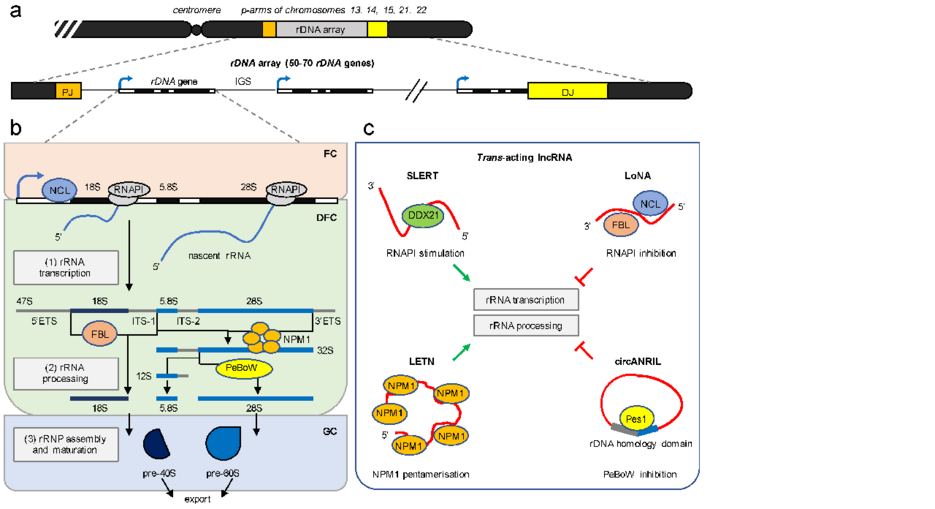
Commuting to work: Nucleolar long non-noding RNA control ribosome biogenesis from near and far
by Victoria Mamontova, Barbara Trifault, Lea Boten and Kaspar Burger
Gene expression is an essential process for cellular growth, proliferation, and differentiation. The transcription of protein-coding genes and non-coding loci depends on RNA polymerases. Interestingly, numerous loci encode long non-coding (lnc)RNA transcripts that are transcribed by RNA polymerase II (RNAPII) and fine-tune the RNA metabolism. The nucleolus is a prime example of how different lncRNA species concomitantly regulate gene expression by facilitating the production and processing of ribosomal (r)RNA for ribosome biogenesis. In the review the authors summarise the current findings on how RNAPII influences nucleolar structure and function. They describe how RNAPII-dependent lncRNA can both promote nucleolar integrity and inhibit ribosomal (r)RNA synthesis by modulating the availability of rRNA synthesis factors in trans. Surprisingly, some lncRNA transcripts can directly originate from nucleolar loci and function in cis. The nucleolar intergenic spacer (IGS), for example, encodes nucleolar transcripts that counteract spurious rRNA synthesis in unperturbed cells. In response to DNA damage, RNAPII-dependent lncRNA originates directly at broken ribosomal (r)DNA loci and is processed into small ncRNA, possibly to modulate DNA repair. Thus, lncRNA-mediated regulation of nucleolar biology occurs by several modes of action and is more direct than anticipated, pointing to an intimate crosstalk of RNA metabolic events.
The full text of the review can be found here (Non-Coding RNA).