Single cell transcriptomics
For single cell analysis we offer two different platforms, 10x Genomics Microfluidics (> 10,000 cells) or FACS-sorted single cell analysis using the SMART-Seq v4 Ultra Low Input protocol (1-1000 cells). This allows analyses such as single cell gene expression, single cell immunoprofiling and ATAC (Assay for Transposase Accessible Chromatin) to be performed.
The protocols offered (10x genomics) are as follows:
- 3' whole transcriptome gene expression
- Identification cell types
- Determination of the heterogeneity
- Finding new cell-surface markers or other cell-specific targets
- Assessment of immune activation as a result of adding a compound to cells
- Comparsion of sample before and after treatment at high resolution
- Detection of immune cell clonality, diversity, antigen specificity, and cellular context
- 5’ whole transcriptome gene expression
- Characterization of individual T-cells and B-cells
- Identification of V(D)J gene sequences
- Pairing of α and β chain TCR sequences from individual T-cells
- Pairing of heavy and light chain immunoglobulin sequences from individual B-cells
- Simultaneous measurement of TCR, B cell Ig, cell surface protein expression, antigen specificity, and 5’ gene expression
- Investigation of open chromatin profiles of individual nuclei
- Analysis of hundreds to thousands of individual nuclei in every run
- Enables the discovery of regulatory motifs and provides a deeper understanding of gene regulation
- Simultaneous profiling of gene expression and open chromatin from the same cell
- Fixation of samples (human, mouse) at the time of collection to lock in their biological states and preserve fragile cells
- Reducing experimental variability and increasing efficiency by batching and multiplexing samples
- Simultaneous performance of highly sensitive measurements of gene expression and cell surface proteins
- Allows biobank samples or samples that cannot be processed fresh to be used for single cell sequencing
- Isolation of nuclei is necessary to obtain additional layers of cellular information, such as chromatin accessibility
Examples of publications:
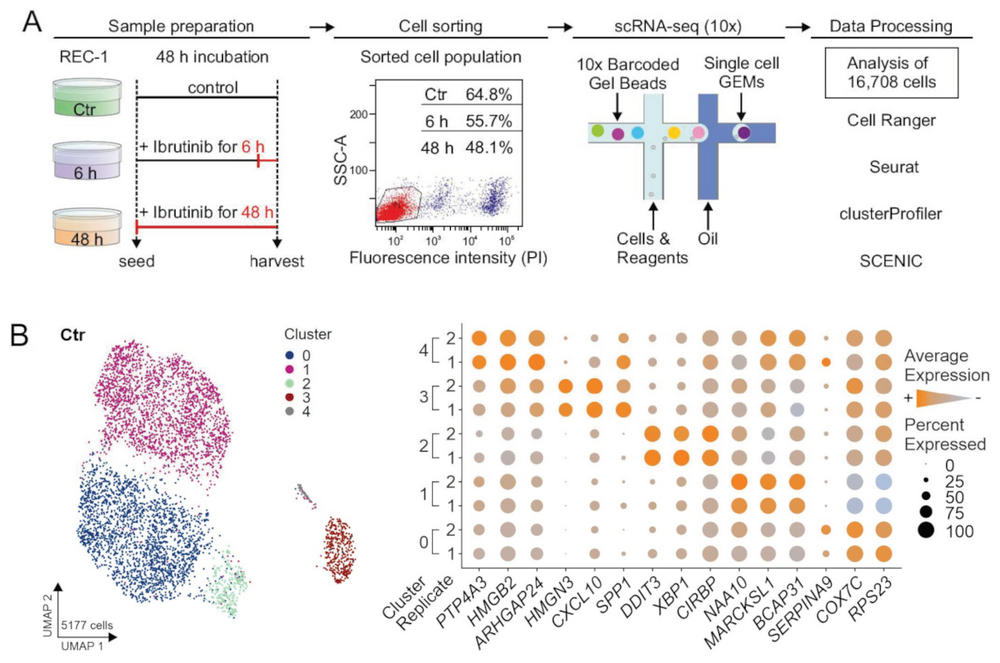
Fuhr V, Vafadarnejad E, Dietrich O, et al. Time-Resolved scRNA-Seq Tracks the Adaptation of a Sensitive MCL Cell Line to Ibrutinib Treatment. IJMS. 2021;22(5):2276. https://doi.org/10.3390/ijms22052276
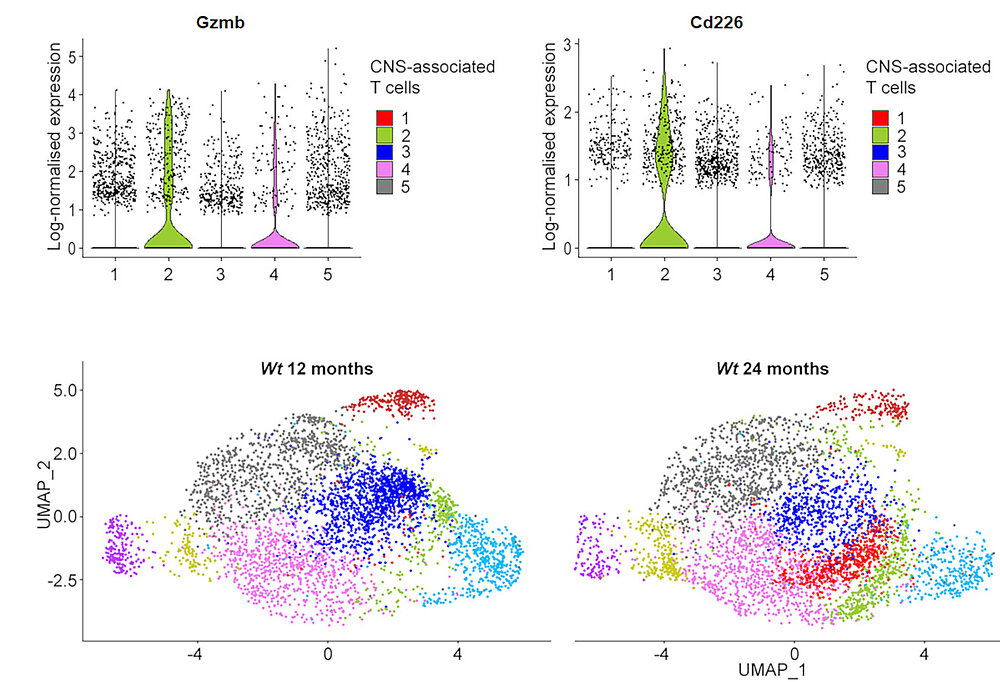
Abdelwahab T, Stadler D, Knöpper K, et al. Cytotoxic CNS-associated T cells drive axon degeneration by targeting perturbed oligodendrocytes in PLP1 mutant mice. iScience. 2023;26(5):106698. https://doi.org/10.1016/j.isci.2023.106698