The VRC Project
The Virtual Research Campus (VRC) is a modern data-management platform currently being developed in collaboration with Bioinformatics Würzburg (Prof. Thomas Dandekar), the Service Center for Medical Informatics (SMI; University Hospital Würzburg), and the Core Unit for Research Data Management. Inspired by successful VRE models (e.g., Charité VRE, EBRAINS project, CERN VRE), our VRC is designed to help researchers store, share, and discover data; to provide secure access; and to foster innovative use and collaborative work.
As a comprehensive data-management solution, the VRC supports research and medical projects in Würzburg and affiliated (cross-site) research communities. It enables the storage, retrieval, analysis, and exchange of data, and offers:
- Metadata management (automatic capture and assignment of descriptive information)
- Versioning & provenance (change tracking to ensure reproducibility)
- User & access-rights management (flexible roles-and-groups model to protect sensitive information)
- Compute-resource & analysis-tool integration (connections to HPC clusters, container environments, and web services)
- Workflows & pipelines (automation of complex analyses via graphical or script-based interfaces)
- Interoperability (APIs and interfaces to common databases and standards)
- Search & data catalogues (full-text search, filters, centralized overviews)
- Documentation & guidelines (embedded wikis, templates, best-practice examples)
- Audit trails & logging (comprehensive recording of all accesses and activities)
By removing traditional barriers, the VRC accelerates innovation—for example, by simplifying data exchange between university hospitals and academic institutions. To make the platform suitable for sensitive clinical data, we plan to implement a fully GDPR-compliant system. Remaining challenges include aligning technical measures (encryption, logging, access controls) and organizational processes (data-processing agreements, user training, risk assessments) with the requirements of the European General Data Protection Regulation. Our aim is to offer commissioned data-processing services for health-related research projects in full adherence to GDPR.
Collaboration with partners such as the NFDI network, SMI, Charité VRE, Bioinformatics Würzburg, the university computing center, and other universities and clinics ensures that our platform meets the latest standards in data management and security.
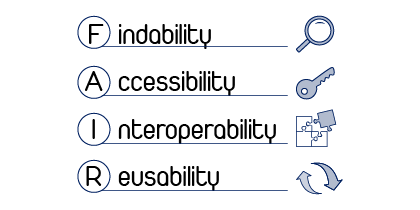
The VRC is firmly grounded in the FAIR principles—Findability, Accessibility, Interoperability, and Reusability. It promotes interoperability with international data communities and aligns us with the missions of the Bavarian Health Cloud and the European Open Science Cloud (EOSC).
FAIR core functions of the VRC:
- Findable:
- Integrated capture, indexing, and storage of metadata
- Support for persistent identifiers and flexible data organization
- Accessible:
- Transparent access policies
- Robust authentication and authorization procedures
- Interoperable:
- Seamless integration with international data communities
- Support for common data standards
- Reusable:
- Annotation tools for rich metadata descriptions
- Internal provenance tracking
- Datalad support for tracking the origins of code and metadata
Architecture of the Virtual Research Campus (VRC): From the Researchers’ Perspective
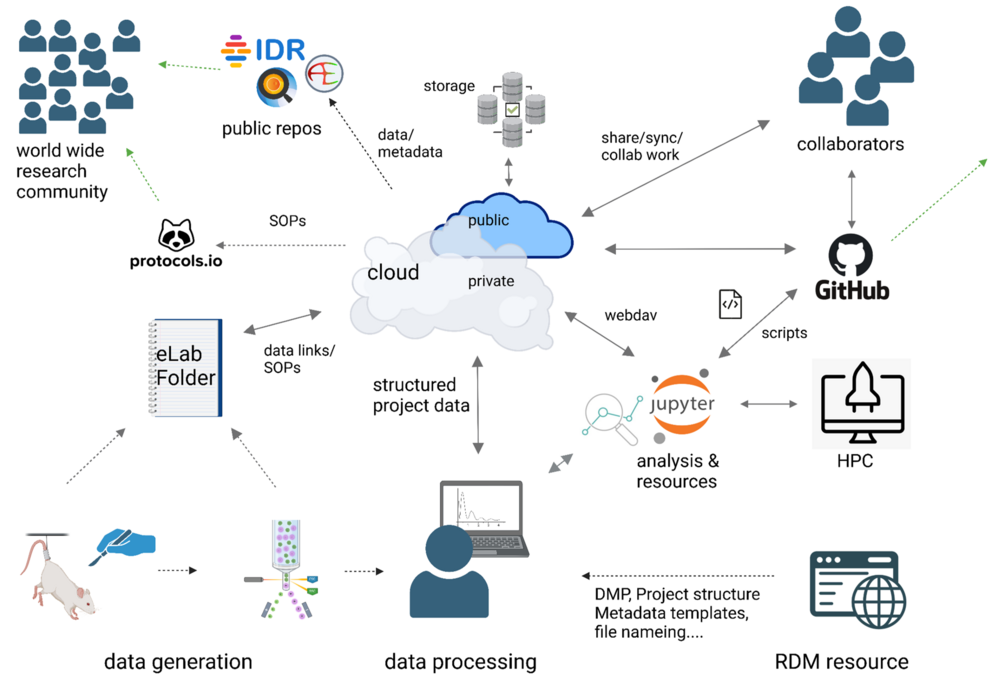
Here’s how your workflow in the University of Würzburg’s VRC works in practice:
- Data Generation & Documentation
- Raw data are produced in the lab (animal experiments, cell culture, measurement instruments).
- Documentation is managed via the electronic lab notebook (eLabFolder): SOPs, protocols, parameters, and links to every data file are stored centrally.
- Metadata and analysis scripts are created in parallel—so all of your information and code (Python, R, MATLAB, etc.) stay together.
- RDM Knowledge Base & Standards
- In the central RDM portal, you’ll find templates for folder structures, file names, and metadata (aligned with NFDI/EU guidelines).
- FAIR checklists indicate recommended formats and versioning tools (Git, Zenodo, Nextcloud).
- Organized Locally or in the Cloud
- Create your project folders using the standard template (RawData, ProcessedData, Metadata, Analysis, Documentation).
- You can work locally just as easily—as long as everything remains uniform and follows the guidelines.
- Cloud Sync & Data Protection
- A WebDAV connection automatically mirrors your folders to the private cloud.
- Automatic backups, snapshots, and version-control–style mechanisms guard against data loss.
- GitHub for Scripts & Collaboration
- Develop your analysis scripts directly in a GitHub repo.
- Versioning, pull requests, and issue tracking simplify teamwork and ensure full provenance.
- FAIR Publication
- With a single click, export selected datasets and metadata to public repositories (e.g. IDR)—complete with DOI, open licenses, and full metadata.
- Your research becomes globally reusable under FAIR principles.
Your Benefits
- Time Savings: Fewer manual imports and format conversions
- Security: Comprehensive protection for your sensitive data
- Reproducibility: Every analysis can be recreated exactly, at any time
- Flexibility: Use your familiar tools on a scalable platform
- Transparency: Metadata and protocols give full insight into your research pipeline and facilitate discovery and integration with other datasets.
Future Architecture of the VRC/VRE: Seamless Integration & State-of-the-Art Orchestration
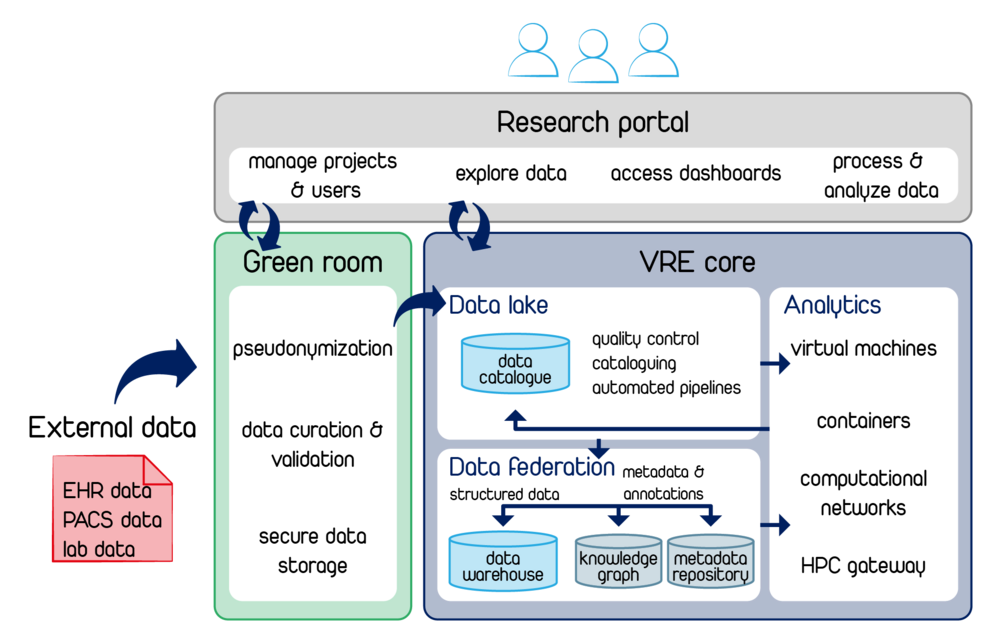
The future platform is built on a fully containerized Kubernetes environment, where all services work together seamlessly and efficiently. This creates a scalable, fault-tolerant research infrastructure that covers the entire data life cycle.
- Modular Web Portal
- Why? Unified access control and single sign-on for researchers.
- How? Runs as its own Kubernetes pod with OAuth/SAML authentication; communicates over a service mesh (encrypted) with other microservices.
- “Green Room” for Sensitive Data
- Why? Early pseudonymization and validation to ensure GDPR compliance.
- How? In a dedicated Kubernetes namespace, encryption and validation tools write raw data into a protected storage backend.
- Automated Data Ingestion
- Why? Heterogeneous clinical and research data must be consistently consolidated.
- How? Multi-stage ingestion pipelines (Connector → Harmonizer → Ingestion Service) move data from EHR/PACS/laboratory systems into project-specific data warehouses and automatically populate the metadata catalogs.
- Project-Specific Data Warehouses & Knowledge Graphs
- Why? Isolation of research cases and semantic querying across entities.
- How? Each project gets its own data warehouse (e.g. PostgreSQL) and a knowledge-graph container (e.g. Neo4j). Metadata services update ontologies and annotations in real time.
- Containerized Analysis Framework
- Why? Reproducible, scalable workflows for deep learning, simulations, and standard analyses.
- How? On-demand pods (JupyterLab, Nextflow) mount data warehouses via CSI volumes. An HPC gateway automatically routes compute-intensive jobs to the cluster.
- FAIR & Metadata Management
- Why? Findability, interoperability, and reusability must be guaranteed throughout.
- How? Persistent identifiers and up-to-date metadata catalogs enable interoperability and reusability. Provenance logs and versioning record every step.
- Kubernetes Orchestration
- Why? Automated self-healing, rolling updates, and horizontal scaling ensure availability and performance.
- How? Helm charts define configurations; the service mesh secures traffic; event streaming loosely couples microservices.
- Seamless Collaboration & Publication
- Why? Research results should be shared efficiently and made visible worldwide.
- How? Real-time collaboration via web interface/WebDAV/CSI volumes: team members can work simultaneously on the same data and documents. Approved datasets are automatically published to repositories (Zenodo, IDR) with DOI and license management.
Summary:
All components—from data capture in the Green Room, through structured warehouses and containerized analysis workspaces, to automated publication—are logically and efficiently connected. Kubernetes orchestrates the entire platform so it remains scalable, secure, and FAIR-compliant.
Why this architecture is crucial:
- Logical, Efficient Orchestration: Kubernetes and a service mesh let each microservice start, communicate, and fail independently without impacting the whole system.
- Seamless Data & Metadata Flows: Automated pipelines link data sources, curation services, and analysis workspaces end-to-end—eliminating redundancy and manual imports.
- High Scalability & Fault Tolerance: Container-based self-healing, horizontal scaling, and canary deployments ensure the system dynamically adapts to growing user loads or resource demands.
- Reproducibility & FAIR Compliance: Versioned data, complete provenance logs, and persistent identifiers guarantee that research outputs remain traceable and reusable indefinitely.